Genomic MRI is a set of tools for analyzing short and mid-range inhomogeneity in nucleotide sequences. Genomic MRI allows the user to upload a nucleotide sequence of their choosing into a saved session which a user may later resume for convenience. [Sessions last 2 weeks.] Short-range inhomogeneity is analyzed by SRI Analyzer, which creates an oligonucleotide composition file for the chosen nucleotide sequence. SRI Generator creates randomized sequence files with the composition specified by any oligonucleotide composition file. Mid-range inhomogeneity is analyzed by MRI Analyzer, which creates MRI composition files and graphs for a set of user-specified parameters (content type, window size, and upper and lower thresholds). MRI Generator creates randomized sequences that mimic the mid-range inhomogeneity in a particular MRI composition file while preserving short-range inhomogeneity. CDS Generator creates randomized sequences that preserve the original protein sequence and di-codon bias. Download Files allows users to conveniently download any file from the above analyses.
One may start or resume a session by using the link on the homepage, the top navigation bar, or by following this link.
Starting a Session
- One can paste in a FASTA-formatted nucleotide sequence file into the text area. We do not recommend this method as the size of sequence may be too large for the input buffer. Instead use method (b) to upload a file as described below.
- To start a session, click on the "choose file" button and browse your local computer for any FASTA-formatted nucleotide sequence files.
- Once you have selected a file, hit the "Start new session with this file" button to upload your file. Depending on the network traffic or the size of your file, the upload process may take a few seconds to a few minutes. The maximum file upload size is 8M.
- Upon successful file upload, the web site will give you a "session label" (six letters, alphanumeric, case-sensitive). You will need this session label in order to resume your current session at a later date (no more than 2 weeks). Please make a note of it.
- Your uploaded nucleotide file will be selectable in SRI Analyzer and MRI Analyzer. We shall refer to this file as the "user file" or userfile. You cannot upload more than one userfile per GMRI session.
- To resume a session, type in your six-character, alphanumeric, case-sensitive session label into the text box marked "Enter a session label".
- Hit the "Resume session" button. If successful, all of your previously saved files and data will be available. You will be automatically forwarded to the last page you worked on.
- If you are unsuccessful, please make sure you are typing in your session label with the appropriate case. Moreover, sessions expire after 2 weeks.
SRI Analyzer is used to analyze the short-range inhomogeneity of FASTA-formatted nucleotide sequences. It outputs an oligonucleotide composition file and a table of most and least common oligonucleotides.
Input
- The user may select any nucleotide sequence files from the listbox marked "File to analyze". These must be FASTA-formatted sequences. They may be uploaded or generated sequences.
- The user must specify the maximum oligomer size to be analyzed from the listbox; the allowed values are 1 to 9. For example, if "3" is selected, the composition will contain the frequencies of all 1-mers, 2-mers, and 3-mers for that nucleotide sequence.
- To analyze the nucleotide sequence file, click the "Analyze File" button when all parameters have been set to satisfaction.
Output
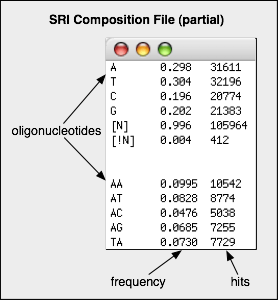
- SRI Analyzer produces a composition file with the oligomers sorted by their size in ascending order (1-mer, 2-mer, etc.).
- SRI Analyzer also produces a composition table with the least and most common oligonucleotides found for each oligomer level.
- The user may download said files by clicking on the links.
- The composition file is saved with a default extension of "
.comp.txt".
- The composition table is saved with a default extension of "
.tbl.txt".
- To preserve proper linebreaks, Windows users should open these files with WordPad, NOT Notepad. MAC users may use TextEdit.
SRI Generator creates randomized FASTA-formatted nucleotide sequences with the same oligonucleotide composition as a source composition file at a specified oligomer level, while following the sequence layout of a specified nucleotide sequence file. In short, SRI Generator creates random sequences that preserve the short-range inhomogeneity of the specified sources.
Input
- The user may select any of the nucleotide sequence files from the listbox marked "Source sequence file". The generated file(s) will have the same length as this file.
- The user next selects a composition file from the listbox marked "Source composition file". These are the composition files generated by SRI Analyzer. Currently, there is no method to upload custom composition files, though composition files of SRI-analyzed randomized sequences may be selected.
- The user may choose to generate more than one random file (up to 30) by selecting from the listbox marked "Number of samples to generate".
- The user must specify the oligomer level (oligomer size) from the table by checking the radio button of the desired oligomer level. The generated file will have a nucleotide composition the same as the source nucleotide composition file up to and including the checked level. In other words, the frequencies of each oligomer will be maintained up to that level.
- Hit the "Generate File" button to generate the randomized file. If the user generates a file twice with identical parameters, the previously generated file shall be given. To generate a new file with the same parameters, simply increase the "Number of samples to generate".
Output
- SRI Generator outputs randomized FASTA-formatted nucleotide sequence file(s) with an oligonucleotide composition as specified by the input parameters. The user may download said file(s) by clicking on the provided link(s).
- The file(s) will have the same name as the source nucleotide sequence file, but with a suffix of "
.rand##_#", where "##" is the file number (1 to 30 possible) and where "#" is the oligomer level.
MRI Analyzer analyzes the mid-range inhomogeneity of a specified nucleotide sequence file by scanning for "rich" and "poor" regions of a particular nucleotide content type using the user-specified window size and upper/lower thresholds. The output is displayed graphically and textually.
Input
- The user must select a nucleotide sequence file from the listbox marked "File to analyze". This is the file to be scanned for mid-range inhomogeneity.
- The listbox marked "Content type" determines what content type a region of a sequence is "rich" in, that is, where a region is abundant in a particular nucleotides. Content types include the single nucleotides A, C, G, T and nucleotide couples AG, GC, GT. Observe here that for nucleotide couples, "GC" refers to 'G' or 'C', not the subsequence "GC". Because each content type also includes its complement, these seven content types cover all possible combinations of nucleotides (eg: "GC" is merely the inverse of "AT").
- The user must enter a positive integer value into the text box marked "Window size" to specify the length of the region for which a subsequence may be considered rich for a particular content type. Currently the analysis uses non-overlapping windows. The default is 50.
- The listbox after the threshold parameters contains the values "by nucleotide" and "by percentage" which govern what type of input is appropriate for the thresholds. A percentage may be given as a whole number or decimal format. For specifying nucleotide thresholds, the upper threshold must be less than or equal to the window size. The lower threshold must be greater than or equal to 0.
- The text box marked "Upper threshold" specifies the percentage or number of nucleotides needed to be considered rich for the specified content type in the specified window size.
- The text box marked "Lower threshold" specifies the percentage or number of nucleotides needed to be considered poor for the specified content type in the specified window size. The complement of the specified content type (i.e., if "A" is specified as content type the complement would be "GCT") would then be considered rich by mutual exclusion.
- Hit the "Analyze File" button to execute the program. If the program fails to execute, try clicking the button twice in a row to ensure all parameters have been received. (Each parameter must be updated internally through a page reload after each change in value. Using the Tab key to exit a field after setting or changing its value ensures that this takes place.)
Output
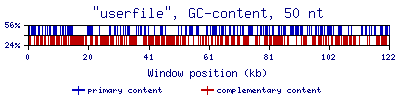
- MRI Analyzer outputs a graphical display with the primary content type rich regions in blue and the complement rich regions in red. The user may save the graphical file by right-clicking on the image and selecting "Save Image". The user must rename the file with the "
.png" extension. HOWEVER, the user is strongly advised to use Download Files instead, as this will provide a suitable filename and extension.
- The textual output is given as an MRI composition file with the same name as the input nucleotide sequence file but with a suffix in the following pattern: "
.??_###_##..#", where "??" is the content type, where "###" is the window size, where "##" is the upper threshold and "#" the lower threshold specified by the user. An example suffix would be ".GC_50_28..15".
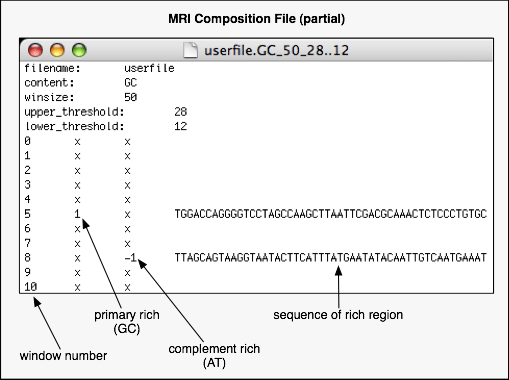
- To obtain the starting and ending indices (let our sequence be 1 to N) for the nucleotide positions of a given window, we may use the window number:
start_index = window_size * window_number + 1 and end_index = start_index + window_size - 1.
MRI Generator creates randomized FASTA-formatted nucleotide sequences with the same oligonucleotide composition as a source composition file, but also mimics the mid-range inhomogeneity of a source MRI composition file.
Input
- The user must select an MRI composition file (they are the ones created by MRI Analyzer) from the listbox marked "MRI composition file to mimic". This file is used to mimic the mid-range inhomogeneity.
- The user next selects an oligonucleotide composition file (they are the ones created by SRI Analyzer) from the list box marked "Composition file to use". This file allows the short-range inhomogeneity to be preserved.
- The user must select an oligomer level (oligomer size) by checking a radio button next to the desired oligomer level in the table. The randomized file created will have the same oligonucleotide composition up to and including the specified oligomer level.
- Hit the "Generate File" button to execute the program.
Output
- MRI Generator creates a randomized FASTA-formatted nucleotide sequence file with the same oligonucleotide composition as the input composition file, up to the specified oligomer level, and with mid-range inhomogeneity characteristics that mimic the specified MRI composition file. This file is known as a "mimic file" or just "mimic".
- To save the mimic, click on the provided link. (Saves usually default to the user desktop.) The file will contain the name of the MRI composition file being used, but with a new suffix in the pattern: "
_mimic#", where "#" is the oligomer level specified by the user.
CDS Generator creates randomized FASTA-formatted nucleotide sequence files that preserve the protein sequence and di-codon bias.
Input
- The user must specify a dicodon frequency table form the listbox marked "Dicodon Frequency Table". The default species is "Human". Other species will be supported in the future.
- The user next selects sequence file type from the listbox marked "Sequence Type". The default sequence file type is FASTA-formatted protein sequences. These are amino acid sequences. In the future FASTA-formatted nucleotide sequence files will also be supported.
- The user may cut and paste in a FASTA-formatted sequence of the specified type into the text area marked "Input FASTA sequence". Alternatively, the user may click on the "Choose file"/"Browse..." button and select a FASTA-formatted file containing sequences of the specified type from the user's local computer. If both the text area and file parameters are supplied, the file upload takes precedence over the text area.
- If you are using PubMed's NCBI Sequence Viewer v2.0 to obtain sequences, then on their Viewer web page you may click on the "Display" list box and select "FASTA" to get a print out of your selected sequence in FASTA format.
- Clicking the "Generate Randomized File" will make a randomized nucleotide sequence appear in a text area below. Clicking the button again with the same sequence in the text area will generate a new randomized sequence.
Output
- A randomized FASTA-formatted nucleotide sequence is provided in a text area marked "Generated sequence". The nucleotide sequence preserves the protein sequence and dicodon bias.
- The user may click in the text area, select all, copy, and then paste the randomized sequence into a document to the save the sequence as a file.
- CDS Generator is currently offered as a stand-alone utility and does not belong to the user's GMRI session (no file integration or resume features).
Download Files
Download Files provides a categorical view of all files generated or uploaded by the user for the given session. The categories include nucleotide sequence files (including both uploaded and generated), oligonucleotide composition files, composition table files, MRI composition files, and image/graph files. To download a file, simply click on the link provided. The browser should prompt the user for a location or default to the user's desktop. Non-image files have a default ".txt" extension appended to their names for convenience.
Download Files also provides a link at the top of the page that allows the user to download all files from the current GMRI session (excluding the original nucleotide sequence file) in a single archive. The archive is a gzipped 'tar' archive (a.k.a. "tarball") with a ".tar.gz" extension and can be opened by most archive file handlers. Be aware that the size of the archive can be quite large if you have generated multiple nucleotide sequence files or MRI composition files with many spikes.


